Abstract
Background CLL incidence in Asians including Chinese is 10 to 20-fold less compared with persons of European descent. We hypothesized an endemic infection in Asia could have been a selection pressure coincidentally resulting in decreased susceptibility and/or resistance to developing CLL in early humans migrating from Africa to Asia about 60-80 KYA. A genome-wide association study (GWAS) study reported 13 risk SNPs involving ACTA2-FAS; TERT; CDKN2B-AS1; ODF1-KLF10; C11orf21, TSPAN3; PMAIP1; BMF; QPCT-PRKD3; BCL2; ACOXL-BCL2L1; LEF; CASP10-CASP60-808 were associated CLL risk in Europeans. We studied these SNPs in 4 populations: Chinese with and without CLL and Europeans with and without CLL.
Methods Genomic DNA was extracted from blood mononuclear cells from 194 consecutive Chinese subjects with CLL diagnosed according to iwCLL 2008 criteria and randomly divided into discovery (N = 93) and validation (N = 101) cohorts. Genomic data from Chinese from the 1000G genome and from Europeans from data of Slager et al. were interrogated. Genotyping was performed by Sanger sequencing, Hardy-Weinberg equilibrium tests (HWE) were done for each SNP and association analyses done with an additive model. Frequencies were compared by Chi-square test adjusted for multiple testing. P < 0.05 was considered significant.
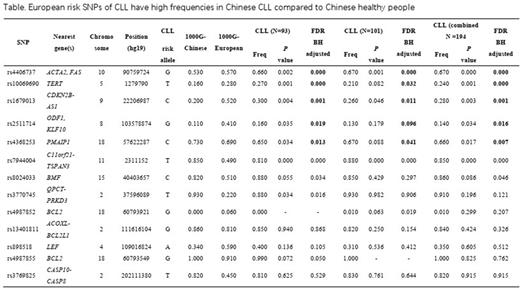
Results The 13 CLL risk SNPs had a heterozygous polymorphism distribution in Chinese with CLL. In the discovery cohort we found allele frequencies of TERT (rs10069690); ACTA-FAS (rs4406737); CDKN2B-AS1 (rs1679013); ODF1-KLF10 (rs2511714) were significantly lower in Chinese without CLL compared with Europeans without CLL but higher in Chinese with CLL compared with Chinese without CLL These findings were confirmed in the validation cohort (Table). rs4368253 SNP in PMAIP1 had a lower allele frequency in Chinese with CLL compared to normal Chinese (0.66 vs. 0.73, P = 0.02; Table).
Conclusions Our data indicate SNPs of TERT (rs10069690); ACTA-FAS (rs4406737); CDKN2B-AS1 (rs1679013); ODF1-KLF10 (rs2511714) are associated with increased CLL risk in Chinese and in Europeans. The decreased frequency of these SNPs in the Chinese population may explain decreased CLL risk in Asians. In contrast, the genetic loci rs4368253 in PMAIP1 may protect Chinese from CLL. Validation of our findings is needed.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal